Macrophages are a key element of the both the innate and adaptive immune response to TB and they can differentiate into M1 and M2 phenotypes based on the cytokines present within their microenvironment. Polarization, particularly to the M1 macrophage phenotype, however, has been looked at more as driving better microbicidal capacity. The authors believe that this may be driven by M1 macrophages possibly expressing innate immunity regulatory genes and gene clusters (Inregs) and therefore sought to delve more into the transcriptional profile of both M1 and M2 macrophages in order to uncover this.
An examination of human peripheral blood and cord blood-derived macrophages, polarized to M1 and M2 phenotypes, showed that they differentially regulate the growth of intracellular Mtb through nitric oxide and autophagy (Figure 1). Differing levels of nitric oxide, reductive oxidative species and autophagic sorting of mycobacteria were taken to account for the Mtb-inhibitive state in M1 macrophages versus Mtb-permissive states in M2 macrophages.
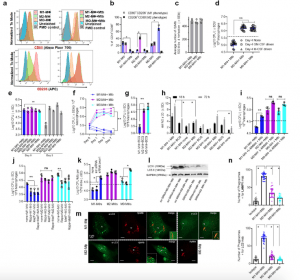
Figure 1: Human umbilical cord and peripheral blood-derived macrophages show heterogeneity in mycobacterial killing associated with oxidants and autophagy. Human cord blood (CBM) or healthy donor PBMC-derived MФs were cultured in the presence of either recombinant human IFN-γ (M1; 10 ng/mL) or human IL-4 (M2; 10 ng/mL) for 5 days and rested for 2 days. Untreated cells were M0-MФs. a, b Surface expression of receptors by CBM-derived naïve and Mycobacterium tuberculosis (Mtb; H37Rv)-infected M1- (CD80+/CD206−) and M2-MФs (CD80−/CD206+) on day 3 using flow cytometry and quantitation (*p < 0.01 t test); gating strategy shown Supplemental Fig. 1. c CBM-derived differentiated MФs were infected with Mtb for 4 h followed by microscopic counts of rfp-labeled MtbH37Rv to determine uptake verified using CFU counts. d PBMC-derived MФs from five healthy donors were differentiated using the indicated cytokines followed by infection with Mtb and CFU assay on day 4. Each point represents one donor (**p < 0.05; Kruskal–Wallis test). e PBMC-derived MФs were differentiated using GM-CSF (M1), IL-4 (M2a), IL-1β M2b), IL-10 (M2c) or left intreated (M0) followed by Mtb infection and CFU assay on day 4 (**p < 0.007). f CBM-derived, cytokine differentiated MФs were infected with Mtb followed by CFU assay over time (**p < 0.006; data from 3 experiments shown). g CBM-derived, differentiated MФs were infected with M. bovis BCG followed by CFU assay on day 4 (**p < 0.005). h CBM-derived and differentiated uninfected MФs or those infected with Mtb or BCG were incubated and at indicated time points, cultures were tested for nitrite using diaminofluorescein diacetate and fluorometry (*,**p < 0.005, t test). QPCR for mRNA of iNOS and protein are shown in Supplemental Fig. 3a and reactive oxygen species levels in Supplementary Fig. 3b. i MФs infected with Mtb as in panel h were incubated in NMMA (0.5 mM; N-monomethyl l-arginine) followed by CFU assay on day 3 (**p < 0.009). j CBM-derived, differentiated MФs infected with Mtb were incubated with 10 µM Rapamycin, 100 µM Metformin or their combination followed by CFU assay on day 3 (**p < 0.009). k CBM-derived, M1-, M2- and M0-MΦs were treated in the presence or absence of siRNA vs. beclin1 (ATG6) or its scrambled control followed by infection with Mtb and CFU counts on day 3 (*p < 0.007). Blot validation of Knockout using siRNA vs. beclin1 (ATG6) is shown in Fig. 3h. l MΦ lysates of panel k collected at 18 h were analyzed using western blots for the lipidation of microtubule-associated light chain 3 (LC3). Lipidation is indicated by LC3-II. m CBM-derived MΦs were infected with rfpMtbH37Rv and stained for an LC3 autophagy marker or LAMP1 lysosome marker using specific antibodies, Alex-Fluor485 conjugates, and imaged using confocal microscopy. Panels illustrate LC3 colocalization; LAMP1 stains using gfpMtbH37Rv is shown in Supplementary Fig. 3c. n Quantification of phagosomes colocalizing with LC3 are shown using an N90 Nikon fluorescence microscope (IF) and Metaview software (*p < 0.004, t test). For panels (b–c–e–f–g–h–i–b–k–n), p-values were calculated using a one-way ANOVA with Tukey’s post-hoc test; one of 2–3 similar experiments shown. CFU or IL-2 assays had triplicate wells plated per group or donor. Panels (d, g, i–k) horizontal dotted lines indicate day 0 CFU (4 h post-infection CFU). All Mtb CFU experiments used MOI of 1. (Khan, et al., 2022).
Mtb infection was demonstrated to induce the upregulation of more innate immunity regulatory genes in M1 macrophages in comparison to M2 macrophages, and this allowed M1 macrophages degrade Mtb better.
It was also found that Mtb-infected M1-macrophages secreted several proinflammatory cytokines and showed increased gene and protein expression of CC- and CXC-type chemokines in comparison to infected M2 macrophages. These data would suggest that during the formation of granulomas, the dominance of M1 macrophages, along with the presence of a predominantly proinflammatory environment work in concert to establish dynamic recruitment of immune cells which contributes an immune response that more effectively controls TB.
Through the strong upregulation of multiple genes associated with the autophagic sorting to autophagolysosomes (APLs) within M1-infected macrophages, this work highlights one pathway that accounts for many TB-exposed humans being able to resist active TB disease. Some of these genes could ideally have potential to be used as possible targets for vaccine development, diagnostics or even therapeutics.
Journal article: Khan, et al., 2022. Human M1 macrophages express unique innate immune response genes after mycobacterial infection to defend against tuberculosis. Nature Communications Biology.
Summary by Vanessa Muwanga










