Researchers have unveiled a new mechanism of immune evasion used by Haemophilus influenzae; a bacterium notorious for exacerbating respiratory infections (Figure 1). This microorganism has developed a sophisticated mechanism to evade the host immune system, enabling persistent colonisation of the respiratory tract.
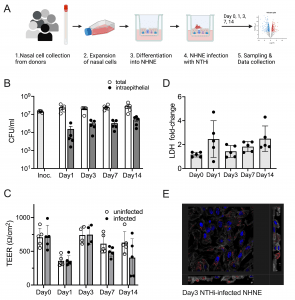
Figure 1: Infection of NHNE from five healthy human donors with H. influenzae strain 86-028NP. Panel A: Schematic representation of experimental workflow. Prepared with BioRender.com. Panel B: Bacterial loads during infection. Total bacterial and intraepithelial bacterial loads (CFU/mL) were determined on Day 1, Day 3, Day 7 and Day 14. Each dot represents the average CFU/mL observed for each donor (n = 2 biological replicates for each donor and condition). ‘Intraepithelial’ bacteria comprise both intracellular and paracellular bacterial populations. Bars and error bars indicate average CFU/mL and standard error. Inoc is the CFU/mL in the bacterial inoculum used to infect NHNE. Panel C: Transepithelial resistance (TEER) changes resulting from NHNE infection. TEER readings for uninfected and infected NHNE for all donors are shown. Due to instrument failure, no readings were available for Donor 1 samples on Day 3 of infection. Average reading and standard error are shown. Panel D: Changes in LDH-release in infected NHNE. LDH levels in infected and uninfected samples were determined using AlphaLISA. The fold change in infected compared to uninfected NHNE from each donor is shown, averages and standard error are shown. Panel E: Immunofluorescence Microscopy of infected NHNE. Representative image showing a Day3 infection of a donor 3 sample. White–Phalloidin stand of filamentous actin; Blue–cellular nuclei, DAPI stain; Green–TUNEL labelling; Red–anti-NTHi antibody. Additional microscopy images for other days of infection are shown in S2 Fig. In Panels A, B &C, data plotted for each timepoint represent the average of multiple biological replicate measurements for samples from an individual donor. https://doi.org/10.1371/journal.ppat.1012282.g001
By inducing a state of immunological tolerance, H. influenzae effectively masks its presence, preventing a robust inflammatory response that would normally eradicate invading pathogens. The bacterium accomplishes this by interfering with the host’s gene expression, specifically downregulating the production of inflammatory molecules.
To elucidate this mechanism, scientists cultured human nasal tissue as a model of the respiratory epithelium. When exposed to live H. influenzae, the tissue exhibited a markedly attenuated inflammatory response compared to the vigorous reaction elicited by heat-killed bacteria. This striking difference underscored the bacterium’s active role in suppressing host immunity.
The ability of H. influenzae to establish chronic infections poses a significant challenge to public health. By understanding the bacterial strategies involved, researchers can develop novel therapeutic approaches to disrupt this pathogen-host equilibrium and restore effective immune function.
Journal article: Kappler, U., et al., 2024. Tolerance to Haemophilus influenzae infection in human epithelial cells: Insights from a primary cell-based model. PLOS Pathogens.
Summary by Stefan Botha










