CTLA4 is a key immune checkpoint involved in immune tolerance and cancer immunotherapy. Patients with CTLA4 deficiency experience immune dysregulation, resembling the adverse effects of immune checkpoint inhibitors used in cancer treatment.
A recent study has unveiled distinct gut microbiome and metabolome signatures associated with CTLA4 deficiency, a genetically acquired immune checkpoint disorder (Figure 1). The study provides new insights into disease mechanisms and potential therapeutic targets. This study represents the first comprehensive analysis of the gut microbiome and metabolome in CTLA4-deficient patients, highlighting potential biomarkers for diagnosis, disease severity, and treatment monitoring.
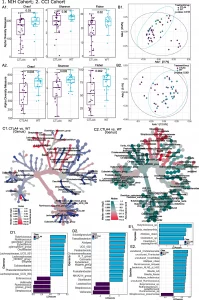
Figure 1: Impact of CTLA4 deficiency on intestinal microbiome signatures. Data for the National Institutes of Health (NIH) Clinical Center and Center for Chronic Immunodeficiency of the Medical Center of the Faculty of Medicine of Freiburg Treatment Consortium (CCI) cohorts are designated by 1 and 2, respectively. Microbiome analyses comparing healthy subjects with patients with CTLA4 deficiency (CTLA4) are shown (NIH cohort: healthy n = 16; CTLA4 n = 32 and CCI cohort: healthy n = 23; CTLA4 n = 23). Alpha diversity measures (Chao1, Shannon, and Fisher) with line and whiskers in the box plot representing the median and inter-quartile range, respectively (A1, A2). Principal coordinates analysis (PCoA) plot of beta diversity based on unweighted UniFrac distances for the two comparison groups with p-values determined by analysis of similarities (ANOSIM) (B1, B2). Heat tree depicting the significant differential abundances of bacterial genera between CTLA4 and healthy (red = higher abundance; blue or teal = lower abundance) (C1 and C2). Linear discriminant analysis (LDA) score determined by the LDA effect size (LEfSe) analysis showing the defining genera (D1, D2) and species (E1, E2) for each cohort. p-values are provided for each comparison with p < 0.05 considered as significant.
To differentiate findings specific to CTLA4 deficiency from general inborn errors of immunity, the study included a control group of patients with common variable immunodeficiency (CVID).
Key Findings: Unique Microbial and Metabolic Profiles
- Microbiome Biomarkers:
- The study identified two bacterial genera that consistently distinguished CTLA4-deficient patients from healthy controls, regardless of geographic origin.
- These biomarkers retained specificity even when compared to the CVID cohort, suggesting their unique association with CTLA4 deficiency rather than general immune dysregulation.
- Metabolome Signatures:
- Metabolomic profiling revealed distinct biochemical pathways associated with gut microbiome changes in CTLA4 deficiency.
- Patients with severe disease exhibited metabolomic profiles distinct from those with milder presentations, correlating with gastrointestinal manifestations and inflammatory markers.
- Therapeutic Implications:
- Immune modulator treatments partially restored microbiome balance, suggesting the reversibility of dysbiosis with targeted therapy.
- These findings point to gut microbial modulation as a potential therapeutic strategy for CTLA4 deficiency and related immune disorders.
This study provides critical insights into the gut microbiome’s role in CTLA4 deficiency, highlighting microbial and metabolic biomarkers associated with disease severity. The findings pave the way for prospective studies to determine the predictive value of these biomarkers and explore their therapeutic potential.
Journal article: Chandrasekaran, P., et al, 2025. The intestinal microbiome and metabolome discern disease severity in cytotoxic T-lymphocyte-associated protein 4 deficiency, Microbiome.
Summary by Stefan Botha










