Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus, emerged in December 2019 in Wuhan, China and is the cause of the coronavirus disease (COVID-19). The virus has overwhelmed healthcare systems and at times put the world on pause as we reflect and find new ways to battle the pandemic.
Currently there seems to be a significant association with COVID-19 and anxiety. The reason being that we do not know how the disease will progress in terms of severity. Several factors, besides clinical risk factors, play a role in disease severity including: diverse immunological, metabolic and muscle abnormalities. As COVID-19 clinically presents in many different ways, there has been a lack of a comprehensive prognostic and explanatory model, all seemingly limited by their use of limited immunological variables, with clinical, radiographic and laboratory features, without the incorporation of a comprehensive assessment of the wide COVID-19 spectrum of abnormalities. In this present study researchers from Mexico, Torres-Ruiz, et al., created and validated a compound explanatory model including diverse clinical, immunological, metabolomic, and muscle atrophy variables to classify COVID-19 patients according to their disease severity and to predict adverse outcomes. In short, their aim was to investigate novel biological features with the hope of explaining COVID-19 severity and prognosis (death and disease progression).
Of the 121 patients recruited, they were divided into groups according to their disease severity as follows:
- Mild/moderate disease: Fever, upper respiratory infection symptoms, with or without pneumonia.
- Severe: Any of the following: respiratory failure, respiratory rate ≥ 30 breaths per minute, oxygen saturation at rest ≤ 93%, PaO2/FiO2 ≤ 300 mmHg.
- Critical: Any of the following: requirement of invasive mechanical ventilation (IMV), shock, multiple organ failure.
The researchers showed that a model comprising Body Mass Index (BMI), hemoglobin, albumin, 3-Hydroxyisovaleric acid, absolute numbers of effector memory CD8+ T cells, T-Helper (Th)-1, Low density granulocytes (LDG)s and the serum/plasma concentrations of monocyte chemoattractant protein 1 (MCP-1), TRIM63, and Neutrophil Extracellular Traps (NETs) is a useful tool for the stratification of patients according to their disease severity. Overall their model displayed good discriminatory capacity and unveiled explanatory features for COVID-19 disease severity as summarised in Figure 1. In addition, inclusive of a distinct metabolic profile, the model may be used as good predictor for disease progression in COVID-19. The model also highlighted novel immunological and metabolomic features associated with COVID-19 severity and prognosis, illustrating interaction between innate and adaptive immunity, inflammation-induced muscle atrophy and hypoxia as the main drivers of COVID-19 severity. This may be the first model that includes clinical features in addition to biological variables that represent varying abnormalities, varying clinical presentations and pathophysiology’s related to disease severity in COVID-19. It must be said that this study does have its limitations including: sample size, genetic background of patients, viral load and viral genotype (associated with differences in disease severity).
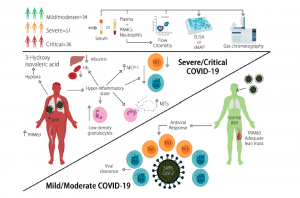
Figure 1: A summary as taken directly from the article: Diverse clinical and biological traits are able to accurately predict COVID-19 severity. Severe/critical disease is characterized by increased BMI, LDGs, circulating NETs, plasma TRIM63, and serum 3-Hydroxyisovaleric acid, as well as diminished levels of Hb, albumin, and lymphopenia of Th1 and CD8+ effector memory T cells (Torres-Ruiz, et al., 2021).
In summary and in their own words,
“The CLICOVID-19 SRS showed the best performance to accurately stratify the COVID-19 severity in both ambulatory and hospitalized patients in an indirect comparison with other predictive indexes…After external validation in different settings, the CLICOVID-19 SRS could be a useful tool to optimize the healthcare resources allocated to manage COVID-19 by identifying subjects that could be safely managed as outpatients and those in whom hospital admission and intensive care is imperative.”
This predictive/risk model is available online as the researchers decided to construct an online risk digital calculator based on the predictive index, which is publicly available (https://rai-unam-mx.shinyapps.io/CLICOVID-19-SRS/)
Journal Article: Torres-Ruiz, et al., 2021. Redefining COVID-19 Severity and Prognosis: The Role of Clinical and Immunobiotypes. Frontiers in Immunology.
Summary by Stefan Botha










