The gut microbiota is an integral component of the human body that is not only important for facilitating digestion and nutrient extraction, but is known to extend wide-ranging impacts on extra-intestinal organ homeostasis and host defense. Recent studies suggest that asthma, a complex, heterogeneous lung disease, is promoted by certain gut microbial populations, however direct evidence implicating a causal effect is lacking, and the underlying mechanisms and cellular players have not been well resolved. In this study, Burrows et al. used animal models and human lung sputum samples to clearly demonstrate that the gut commensal protozoan Tritrichomonas musculis (T.mu) can modulate a subset of intestine-residing group 2 innate lymphoid cell (ILC2) behaviour to remotely shape lung immunity, thereby significantly contributing to the severity and pathophysiology of different lung diseases.
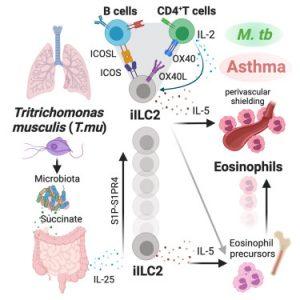
Prior to this study, it has been known that colonization of the gut by commensal Tritrichomonas spp. and pathogens contributes to local ILC2 activation and eosinophil accumulation, features of Type 2 immune defence. However, it is unknown whether T.mu colonization can remotely regulate extra-intestinal organ homeostasis and influence disease outcome. Burrows et al. showed that T. mu gut colonization of mice led to increased trafficking of both eosinophils and gut-derived ILC2s into the lungs. Interestingly, the authors discovered that the accumulation of both these innate immune cells requires a tripartite signaling network involving the T and B adaptive immune cells in the lungs.
To understand the pathological significance following eosinophilia in the lungs, the authors used an allergic asthma disease model. T.mu colonized mice had significantly higher histopathological scores and increased inflammation in the lungs compared to non-colonized. They further showed in a cohort of human patients with severe asthma that there was enrichment of the human-associated gut-resident relatives of murine Tritrichomonas spp DNA sequences in their lung sputum. In contrast, in another lung disease model, the authors showed T. mu colonization allowed accumulated lung perivascular eosinophils to “shield” and prevent systemic dissemination of Mycobacterium tuberculosis. Altogether, these novel findings highlight a roadmap addressing how protozoan-driven gut-lung axis can both drive severity and be protective against different pulmonary diseases.
The roles of how eukaryotic members of the microbiota, particularly commensal protozoa, impact homeostasis and pathophysiology remain underappreciated and understudied. This paper’s findings shed new light and elicit excitement on discovering new mechanisms involving gut protozoans by which we can explore and identify targetable molecular pathways for therapeutic management.
Journal article: A gut commensal protozoan determines respiratory disease outcomes by pulmonary immunity. Cell. 2025
Summary by Zi Yan Chen