The small intestine plays a dual role—absorbing essential nutrients while preventing pathogens from invading the body. This creates a significant challenge for the immune system, as it must effectively detect and eliminate threats without compromising digestion and nutrient absorption.
New research has revealed a surprising adaptation in the immune system: tissue-resident memory CD8 T cells (TRM cells) in the small intestine shift their location to strategically intercept infections before they spread deeper into vulnerable tissues (Figure 1). This study highlights the sophisticated ways in which immune cells adapt to specific organs and offers insights that could advance cancer immunotherapies targeting different tissues.
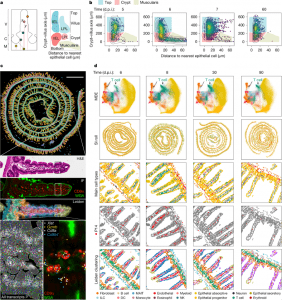
Figure 1: Characterization of the spatial and transcriptional state of antigen-specific CD8 T cells in response to acute viral infection in the mouse SI with spatial transcriptomics. a, Left, a coordinate system to define morphological axes in the SI. C, crypt; M, muscularis; V, villus. Right, the distance to the nearest epithelial cells and the distance to muscularis form the basis of an IMAP. The top of the villus and the crypt regions house both intraepithelial lymphocytes (IEL) and lamina propria lymphocytes (LPL). b, An IMAP representation of P14 T cell localization measured by immunofluorescence (staining at the indicated days after infection. Two biological replicates for n = 3 mice per timepoint, representative data from one mouse are shown. The gates for the top of the villus (blue), the crypt (red) and the muscularis (beige) highlight the different regions. The points, representing cell positions, are coloured by kernel density estimates over the IMAP (x, y) coordinates. c, An overview of the Xenium-based spatial transcriptomics data structure: row 1, Xenium output of a mouse intestine (8 d.p.i.), with cells coloured by Leiden cluster; row 2, a magnification of a villus showing H&E staining; row 3, confocal immunofluorescence images of CD8α and WGA; row 4, Xenium DAPI staining with cell boundary segmentation masks coloured by Leiden cluster; row 5, a further subregion magnification of the same villus depicting Xenium DAPI staining overlaid by cell boundary segmentation and all transcripts assigned to cells (left) and an immunofluorescence image of CD8α and WGA overlaid with transcript locations for Cd8a, Cd8b, Gzmb and female P14-specific Xist transcript locations overlaid (right). d, An overview of the processed Xenium data of mouse SI at 6, 8, 30 and 90 d.p.i. (columns): row 1, cells in a joint minimum distortion embedding (MDE) of all SI Xenium samples coloured by cell type; row 2, in situ spatial positioning of the cells; rows 3–5, magnifications coloured by cell type (row 3), with P14 clusters/cells highlighted in red (row 4) and coloured by Leiden cluster (row 5). One of two biological replicates for each timepoint is shown. DC, dendritic cell; ILC, innate lymphoid cell; MAIT, mucosal-associated invariant T; NK, natural killer cell.
Researchers investigated how TRM cells position themselves to optimize protection. Their findings show that immune cells are not randomly distributed; instead, they follow specific spatial cues from the gut, relocating based on infection risk and immune demands.
Using spatial transcriptomics, a cutting-edge technology that allows researchers to track immune cells within intact tissues, the team mapped TRM cell movements in both human and mouse small intestines.
Their findings identified two distinct populations of TRM cells:
- Progenitor-like TRM cells – These reside in the crypts between the villi (the small, finger-like projections lining the intestine). They act as a reserve force, ready to replenish effector T cells when needed.
- Differentiated TRM cells – These occupy the top of the villi, a more exposed position, where they can swiftly detect and combat invading pathogens before infections spread deeper.
This organization ensures a continuous immune response: while differentiated TRM cells handle immediate threats, progenitor TRM cells remain hidden, prepared to activate if reinforcements are needed.
The study also uncovered how the gut signals immune cells to move. After a viral infection, the small intestine releases specific chemical cues that guide TRM cells to their assigned locations. These signals instruct immune cells on where to go and what to do, reinforcing gut immunity by positioning cells precisely where they are needed most.
Understanding how immune cells adapt to tissue environments could transform cancer immunotherapy. Tissue-resident immune cells may play a critical role in targeting tumors within specific organs, including the lungs, kidneys, and digestive tract.
Journal article: Reina-Campos, M., et al., 2025. Tissue-resident memory CD8 T cell diversity is spatiotemporally imprinted. Nature.
Summary by Stefan Botha










