Recent advancements in tissue engineering have yielded a powerful tool for investigating complex diseases: organoids. These three-dimensional, in vitro cultures recapitulate the cellular architecture and functionality of their in vivo counterparts. In the context of celiac disease, intestinal organoids have emerged as a valuable model system to elucidate disease mechanisms.
A new study leverages intestinal organoids derived from patients with celiac disease to identify a previously unknown molecular link between gluten exposure and intestinal damage (Figure 1). The investigation centred on the role of IL-7, a cytokine implicated in other autoimmune disorders but not previously associated with celiac disease.
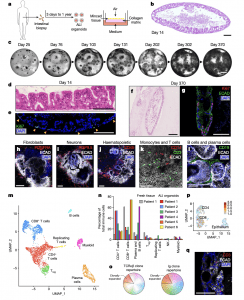
Figure 1: Human ALI small-intestine organoids preserve epithelium, mesenchyme and diverse immune populations without exogenous reconstitution. a, Schematic outlining the steps in creating ALI organoids’. ALI small-intestine organoid with haematoxylin and eosin (H&E) staining at culture day 14 (representative of n = 25 biological replicates). c, ALI organoid growth time course, brightfield (representative of n = 3 biological replicates cultured for more than 300 days). d, Epithelial protrusions at day 14, with H&E staining (representative of n = 25 biological replicates). e, Immunofluorescence staining with Ki67 (green) and DAPI (blue) at day 14 (representative of n = 9 biological replicates). f, ALI organoids at day 370, stained with H&E (representative of n = 3 biological replicates cultured for more than 300 days). g, ALI organoids at day 370 stained with Ki67 (red), ECAD (green) and DAPI (blue) (representative of n = 3 biological replicates cultured for more than 300 days). h–j, ALI organoid whole-mount immunofluorescence staining using ECAD (white) and DAPI (blue) with PDGFRA (red) (h), PGP9.5 (red) (i) or CD45 (red) ( j) at day 14 (representative of n = 3 biological replicates). k,l, Immunofluorescence whole-mount staining of ALI organoids using: ECAD (white), CD14 (red) and CD3 (green) (k); and ECAD (white), DAPI (blue) and CD19 (green) (l), at day 14 (representative of n = 3 biological replicates). m, An scRNA-seq UMAP plot of FACS-sorted CD45 + cells from ALI organoids at day 14, n = 4 donors. n, Organoid immune subset frequencies (n = 6 patients) compared with fresh tissue (n = 1 patient) using scRNA-seq at day 14. o, TCR (n = 6 patients) and BCR (n = 3 patients) repertoire and clonal expansion from ALI organoids at day 14 from scRNA-seq analysis. p, An scRNA- seq UMAP plot (from m) of T-cell CD103 mRNA (n = 4 biological replicates). q, Immunofluorescence staining of CD3 + IELs (red), ECAD (white) and DAPI (blue) on day 14 (representative of n = 3 biological replicates). All figure panels are for duodenal organoids apart from c, f and g, which are ileal. Duodenal and ileal organoids did not exhibit qualitative differences. All scale bars are 100 μm except for c, which is 5 mm.
By examining the cellular responses of these organoids to gluten challenge, researchers have unveiled a complex interplay of immune and epithelial cells. The data suggest a pivotal role for IL-7 in orchestrating the inflammatory cascade that underlies celiac disease pathogenesis. This finding offers a promising therapeutic target for intervention.
The development of organoid technology represents a significant leap forward in celiac disease research. By providing a more physiologically relevant model compared to traditional cell culture or animal studies, organoids enable precise dissection of disease mechanisms and facilitate the evaluation of novel therapeutic strategies.
Journal article: Santos., A.J.M., et al, 2024. A human autoimmune organoid model reveals IL-7 function in coeliac disease, Nature.
Summary by Stefan Botha
