It is almost possible to say that after exhausting efforts from scientists globally, we can effectively manage SARS-CoV-2 with successfully developed vaccines. It is concerning as we move forward, that the current vaccines are based in majority on the early strains of SARS-CoV-2 which emerged in 2019.
The SARS-CoV-2 Omicron variant of concern (VOC) is the variant dominating the headlines and is the major strain out there to date. This strain has presents resistance to the neutralising antibody responses elicited from COVID-19 vaccination (READ MORE). Therefore, there is a need for effective vaccines that may offer a broader range of protection against SARS-CoV-2 VOCs and future VOCs.
In a recent paper, researchers have identified and described an epitope capable of neutralising a variety of coronaviruses including SARS-CoV-1, WIV1 and SHC014. This epitope exists on the receptor binding domain (RBD) of the SARS-CoV-2 S protein. This region, ADG20 (Figure 1), has been found to neutralise SARS-CoV-2 VOCs and may be targeted by a variety of antibodies.
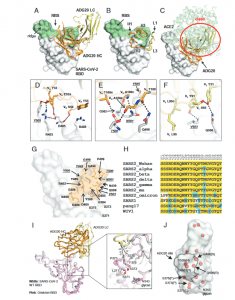
Figure 1: Crystal structure of ADG20 in complex with SARS-CoV-2 RBD. The heavy and light chains of ADG20 are shown in orange and yellow, respectively.SARS-CoV-2 RBD is shown in white. ADG20 epitope residues and RBS residues are defined by residues with surface area buried by ADG20 and ACE2 of>0Å2, respectively, and calculated based on the ADG20/RBD crystal structure in this study and an RBD/ACE2 complex structure (PDB 6M0J) (54) by PISA (50).(A) Structure of ADG20 Fab in complex with SARS-CoV-2 RBD. The RBS is shown in green. For clarity, the constant domains of ADG20 Fab were omitted.(B) ADG20 CDRs that interact with SARS-CoV-2 are shown as tubes. (C) The ACE2/RBD complex structure is superimposed on the ADG20/RBD complex.ADG20 would clash with ACE2 (green) if bound to RBD simultaneously (indicated by red circle). (DtoF) Detailed interactions between ADG20 (D) CDRsH1 and H2, (E) CDR H3, and (F) light chain with SARS-CoV-2 RBD. Hydrogen bonds and salt bridges are represented by black dashed lines. Epitope residuesinvolved in ACE2 binding are underlined and italicized. Kabat numbering is used for antibody residues throughout this paper. (G) Epitope residues of ADG20are highlighted in orange, with side chains shown as sticks and Cαof glycines as spheres. (H) Sequence alignment of RBD residues in a subset of SARS-likeviruses where we also generated PSV neutralization data. Residues within the ADG20 epitope as well as those in the 371 to 375 loop are aligned. Conservedresidues among SARS-related CoVs are highlighted with a yellow background, while similar residues are in a cyan background (amino acids scoring greaterthan or equal to 0 in the BLOSUM62 alignment score matrix (55) were counted as similar here). (I) Structural comparison between the ADG20-bound WTSARS-CoV-2 RBD and Omicron RBD (BA.1 sublineage). Crystal structure of ADG20 (heavy chain: orange, light chain: yellow) in complex with WT SARS-CoV-2RBD (white) is from this study (PDB 7U2D). The superimposed Omicron RBD (pink) is extracted from a previous structure in complex with two other Fabs(PDB 7QNW) (19). (J) The RBD is represented by a transparent white surface. Mutated residues in the Omicron variant (BA.1 sublineage) are shown as redspheres. The ADG20 epitope is highlighted by black solid lines. The following four residues in the ADG20 epitope are mutated in the Omicron variant:G496S, Q498R, N501Y, and Y505 (Yuan, et al., 2022).
Though identification of unique sites in the SARS-CoV-2 RBD, may present an attractive option for combatting sARS-CoV-2 mutation and immunisation escape.
In their own words:
“ADG20 can then benefit from high potency through direct competition with ACE2 in the more variable RBS and interaction with the more highly conserved CR3022 site. Importantly, antibodies that are able to target this site generally neutralize a broad range of VOCs, albeit with reduced potency against Omicron. Thus, this conserved and vulnerable site can be exploited for the design of universal vaccines and therapeutic antibodies.”
Journal article: Yuan, M., et al., 2022. A broad and potent neutralization epitope in SARS-related coronaviruses. Biophysics and Computational Biology.
Summary by Stefan Botha