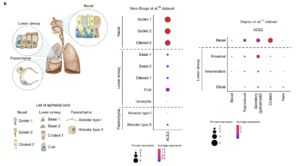
Schematic illustration depicts major anatomical regions in the human respiratory tree demonstrated in this study: nasal, lower airway and lung parenchyma (left). Expression of ACE2 is from airway epithelial cell datasets: Vieira Braga et al.26 (middle) and Deprez et al.27 (right). Datasets were retrieved from existing sources and cell clustering and nomenclature were retained based on the respective studies. For gene expression results in the dot plots, the dot size represents the proportion of cells within the respective cell type expressing the gene and the dot color represents the average gene expression level within the particular cell type. (Source: Sungnak et al., 2020)
Investigators in a brief communication in Nature Medicine utilised the Covid19 cell atlas database to identify gene expression of the SARS-CoV-2 entry receptor ACE2 and other genes potentially associated with pathogenesis using known single-cell RNA sequencing datasets.
The binding affinity of the SARS-CoV-2 Spike (S) protein for ACE2 is known to be a major determinant of viral replication rate and disease severity, and viral entry depends on TMPRSS2 protease activity. Low level expression of ACE2 was found in cells from airways, cornea, oesophagus, ileum, colon, liver, gall bladder, heart, kidney and testis, suggestive that ACE2, and not TMPRSS2, may be a limiting factor for viral entry at the initial stage of infection. The authors went onto investigate nasal epithelial cells in an independent scRNA-seq study from nasal brushings and biopsies and show the highest expression of ACE2 was found in nasal goblet and ciliated cells. The authors conclude that these cells serve as “loci of original infection and possible reservoirs for dissemination within and between individuals.”
These results suggest that viral transmission, and hence the main determining factor of the R0 value, may rely on exploiting existing secretory pathways from nasal goblet cells. This potential mechanism of viral susceptibility and transmission, especially at the pre-symptomatic stage of infection, also indicates that a nasal vaccine/drug strategy should be considered.
Journal Article: Sungnak et al., 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine
Article by Clive Gray










