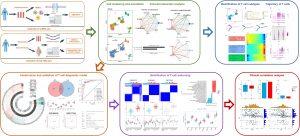
Rheumatoid arthritis (RA) is an autoimmune disease that causes chronic inflammation of the joints, leading to pain, swelling, and joint damage. T cells play a critical role in RA’s progression, making them a key focus for therapeutic intervention. In a ground-breaking study published in Frontiers in Immunology, researchers employed an integrated approach to develop a T-cell-related diagnostic model using single-cell RNA sequencing (scRNA-seq), bulk RNA-seq, Mendelian randomization, and expression quantitative trait loci (eQTL). This study marks a significant advancement in understanding RA’s underlying mechanisms and offers new avenues for personalized immunotherapy.
Study Overview
The research team utilized scRNA-seq to analyze 10,211 individual cells from RA patients, classifying them into seven different subtypes based on marker gene expression. By integrating bulk RNA-seq data and leveraging Mendelian randomization, they identified eight T-cell-related diagnostic features: MIER1, PPP1CB, ICOS, GADD45A, CD3D, SLFN5, PIP4K2A, and IL6ST. These features were used to develop a nomogram model that can accurately classify RA patients into two distinct clusters, based on T-cell involvement.
Using XGBoost machine learning algorithms, the researchers could accurately predict RA progression by identifying pathogenic T-cell marker genes. The two identified clusters, Cluster 1 and Cluster 2, exhibited different immune infiltration patterns, with Cluster 2 showing a higher T-cell score, suggesting a more aggressive disease phenotype.
Key Findings
The most significant finding was the role of T-cell heterogeneity in RA. The researchers found that different T-cell subtypes contributed to varying immune responses in RA patients. The T-cell-related nomogram model developed in this study can serve as a robust tool for diagnosing RA and predicting disease outcomes. Furthermore, ICOS and IL6ST, two critical T-cell markers, were found to negatively correlate with age, suggesting that younger patients might have a more aggressive immune response driven by these markers.
The use of consensus clustering allowed the researchers to identify two unique T-cell patterns in RA. Cluster 2, in particular, was associated with a more inflammatory immune profile, characterized by elevated levels of cytokines and increased T-cell infiltration. This discovery could pave the way for more targeted treatments aimed at modulating T-cell activity in RA.
Implications for RA Treatment
This study has far-reaching implications for the future of RA treatment. By identifying T-cell-related diagnostic features, clinicians can now classify patients into distinct subtypes, allowing for more personalized treatment approaches. This could lead to more effective use of immunotherapies that target specific T-cell subpopulations.
Additionally, the use of machine learning models like XGBoost in predicting RA progression opens new doors for the early diagnosis and intervention of the disease. The nomogram model developed in this study offers a practical, data-driven approach to predicting disease severity and personalizing treatment options for RA patients.
The integrated approach used in this study provides a comprehensive understanding of T-cell heterogeneity in RA. The identification of T-cell-related diagnostic markers and the development of a T-cell-based nomogram model are crucial steps toward personalized RA treatment. Future research should focus on validating these findings in larger cohorts and exploring the potential for T-cell-targeted therapies.
Journal Article: Ding, Qiang, et al. “Integrated Analysis of Single-Cell RNA-Seq, Bulk RNA-Seq, Mendelian Randomization, and EQTL Reveals T Cell-Related Nomogram Model and Subtype Classification in Rheumatoid Arthritis.” Frontiers in Immunology.
Summary by Faith Oluwamakinde