Single-cell RNA sequencing (scRNA-seq) is a laboratory technique that falls under the greater realm of omics technologies that is pushing the boundaries when it comes to Immunology research. As the authors put it; “precise solutions require first the generation of precise knowledge”. In line with this, scRNA-seq technologies are allowing us to decipher more about specific pathological pathways and mechanisms and the particular cell populations responsible for either driving or combating several disease processes so that we can get better at improving healthcare and the lives of others.
Researchers are now in position to decipher more on the innate immune response, by looking into T and B cell receptor repertoires. When it comes to translation in the clinical setting, people have now been able to identify molecular signals that can determine whether patients are responding well to immunotherapy or not. These are aspects that can greatly contribute aid efforts around precision medicine.
With the rise of soo many different technologies and platforms now used to carry out scRNA-seq, one could easily either easily get confused or spoilt for choice as to where to start and what to use. In figure 1, the authors tell us about major platforms currently on the market that have application for Immunology research as well as some factors to consider with regards to the capabilities of each.
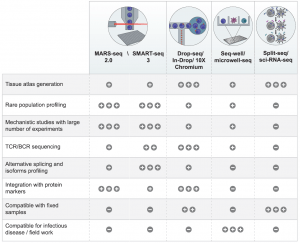
Figure 1: A table summarizing the advantages of different scRNA-seq technologies in immunology research. ‘‘+,’’ ‘‘++,’’ and ‘‘+++’’ indicate the relevant advantage or disadvantage represented by each technology. ‘‘_’’ indicates that the specific technology is lacking the attribute (Ginhoux, et al., 2022).
The authors are frank in highlighting that each platform does have its upsides and pitfalls and that one has to think about the work they are doing and the overall aim one wants to achieve when trying to balance that with the both the strengths and limitations of each platform. This might also be why Ginhoux and colleagues talk about researchers combining more than one platform in their work.
Indeed, with the generation of not only large, but complex datasets that arise from scRNA-seq, there does come a need to have not only intuitive up-to-date algorithms but also multi-layered analysis strategies that can allow one to make sense of the data generated, present it and make scientifically sound conclusions from it. The authors point us to work that gives an in-depth look into analytical challenges that have come up with the rise of single cell genomics technologies and the strategies people have applied to try and overcome them; Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data , Computational Methods for Single-Cell RNA Sequencing, Current best practices in single-cell RNA-seq analysis: a tutorial. These analytical challenges become even more magnified when we look at the shift into single cell multiomics, that involves people combining proteomic, metabolomic and epigenetic datasets in a bid to get a more wholesome picture of the immune system and how it functions.
A phenomenon that has risen with scRNA-seq, and more so with Immunology work, is the constant identification of “novel” and rare cell subsets and subtypes, which have now been found to be identical cell types that simply go under different names in different studies. Where we are now, the development of comprehensive atlases has allowed the integration of data from different datasets, making it easier to identify and characterize similar cell populations across different tissues and body compartments. This is ideally making it easier for people carrying out similar research work better integrate results from different research groups.
With the constant improvement in single cell technologies, and the rise of even more platforms, there is a predicted fall in prices set to occur that could potentially see scRNA-seq become somewhat affordable and even more accessible to researchers globally. When we look at a better future for single cell Immunology, more conversations and efforts around standardized nomenclature and the development of centralized datasets will allow researchers share information more seamlessly and drive more collaborations. The generation of the Human Cell Atlas for example, holds promise for more community-driven efforts that could even see standardization make it easier to interpret data generated across more than one scRNA-seq platform.
Journal article: Ginhoux, F, et al., 2022. Single-cell immunology: Past, present, and future. Immunity.
Summary by Vanessa Mwebaza Muwanga










